Fritz User Guide¶
Fritz is the science data platform for the Zwicky Transient Facility (ZTF) Phase II.
It implements an end-to-end, scalable, API-first system for Time-domain Astronomy, featuring
A multi-survey data archive and alert broker
An interactive collaborative marshal for the transient, variable, and Solar system science cases
A workhorse for machine learning applications and active learning
Follow-up observation management with robotic and classical facilities
Fine-grained access control
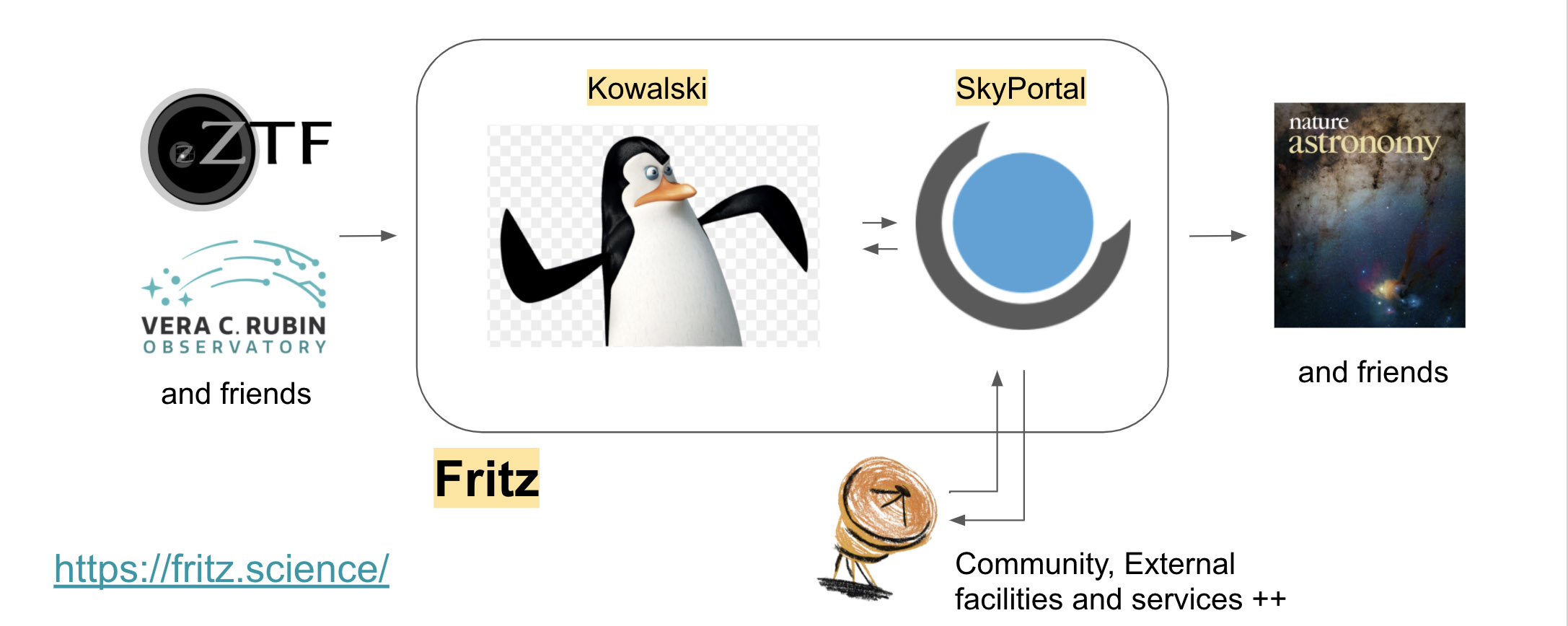
The key characteristics of Fritz are efficiency, scalability, portability, and extensibility. Fritz employs a modular architecture and integrates and extends two major components: Kowalski acts as the alert processor and data archive, and SkyPortal, which handles the rest of the stack. The schematic overview of our system is shown below:
Quick start¶
A quick tour of Fritz from the user perspective¶
User profile¶
Newly invited Fritz users are first taken to the Profile page.
Here, you can update your user info, preferences, and manage tokens to
interact with Fritz programmatically.
If you have a gravatar associated with your email address,
it will be used as your avatar throughout the portal.

System administrators can invite new users to Fritz and manage their roles/ACLs as well as group membership and
alert stream access.
We use OAuth for user authentication.
Dashboard¶
On the Dashboard, the landing page, the user can see a collection of configurable widgets displaying the information on the most popular and latest saved sources, the newsfeed, conditions at telescopes and so on.

Groups and filters¶
New Fritz users are added to one or more groups.
Users can also create groups themselves and request admission to groups they are not a member of.

You can find and manage the list of group members, the sources saved to the group, and the group’s alert filters (defined on one of the alert streams that the group has access to) on the Group page. Group admins can create new filters (and modify existing ones). System administrators can grant alert stream access to the groups.
Fritz provides rich alert stream filtering capabilities through its Kowalski backend,
which consumes the ZTF Kafka alert stream, persisting the alerts to a database,
and supplementing them with additional data such as the Galactic coordinates,
external catalog cross-matches, machine learning scores etc.
Alert filters are implemented as aggregation pipelines and are executed on the enhanced packets containing, in particular, the full photometry history. For a detailed discussion of this, refer to the alert filters section of the docs.
Fritz performs automated checks of the filter definition such that no code audit is necessary. Valid changes are propagated and applied almost immediately. Users can enable/disable filters, inspect filter version history, opt in for automatically saving passing objects to the filter’s group and updating annotations each time an object passes the filter.

Filters can range from very simple that rely on, for example, mostly ML scores, or implement very complicated logic/computations. Watch lists can also be implemented as simple Fritz filters.
Finally, we provide public alert databases for filter design and debugging.
Candidates and Sources¶
Alerts passing a filter are posted to Fritz’s SkyPortal backend as Candidates and appear on the Scanning page (for the corresponding filter groups).
Candidates do not have to originate from Kowalski and could be posted (manually) via the API.
On the Candidates page, the users can filter, scan and inspect the objects that have passed filters of their groups
and save them to one or more groups. Candidates that are not saved to any group within 7 days are removed from Fritz.
Saved Candidates become Sources that are persisted indefinitely.

Candidates/Sources can be supplemented with (structured) annotations, which is particularly useful for scanning:

The Source page aggregates all kinds of information related to an object:

In particular, users can generate finder charts and star lists for the source, check its observability,
add and inspect redshift data:

Annotations provide a way to store structured data related to the source, while comments can be used to enter text/data in free-form. Sources can be classified according to a taxonomy (which users can post themselves provided they have the necessary ACL). Interactive offset plot displays positional information.
Users can send notifications about the source to the group members, provided they opted in for receiving such.

For sources annotated with Gaia color information, an
HR diagram is rendered.

Users can interactively inspect (optionally binned) photometry data in both magnitude and flux spaces.
Tools for data import, export, and granular access management are available.

The photometry plot for sources annotated with period data additionally displays phase-folded light curves:

Tooling for extensive in-browser periodogram analysis for variable sources is also available:

Users can inspect, analyze, and manage (including export/import) spectroscopy data.
We provide interfaces to work with both robotic and classical follow-up facilities, including a framework for dealing with allocations. In particular, we feature automated integration with the SEDM.

Sources can be added to (“classical”) observing runs:

Sources can be added to favorites:

Users can query the objects that exist in Fritz’s SkyPortal on the Sources page:

Only the objects that have been posted to Fritz’s SkyPortal backend are saved in its database.
However, Fritz’s users can access the entire archive of ZTF alerts (~390M as of June 2021) via the Alerts page:

Fritz’s users also have access to the entire archive of photometric light curves of ZTF sources
(~4B as of June 2021) via the Archive page:

Mobile experience¶
Fritz’s interfaces are mobile-friendly, so the app will work as expected on your phone or tablet:
 |
 |
Using the API¶
An API enables access to most of the underlying functionality of Fritz/SkyPortal/Kowalski. The workflows described above are all enabled by specific API calls. The complete OpenAPI specification is available at https://docs.fritz.science/api.html.
To use the API, you will need a token that can be generated on the Profile page. Once you have that, you can access Fritz programmatically as follows.
import requests
from typing import Mapping, Optional
import urllib.parse
token = 'ea70a5f0-b321-43c6-96a1-b2de225e0339'
def api(
method: str,
endpoint: str,
data: Optional[Mapping] = None,
base_url: str = "https://fritz.science",
):
headers = {"Authorization": f"token {token}"}
response = requests.request(
method.upper(),
urllib.parse.urljoin(base_url, endpoint),
json=data,
headers=headers,
)
return response
For example, here is how to retrieve object’s annotations:
object_id = "ZTF21aatjavc"
response = api("get", f"api/sources/{object_id}/annotations")
data = response.json().get("data", None)
print(data)
Which would yield something similar to:
[{
"author_id": 3,
"created_at": "2021-04-08T06:17:56.980090",
"data": {
"acai_b": 0.00011,
"acai_h": 0.98887,
"acai_n": 0.00361,
"acai_o": 0.00925,
"acai_v": 2e-05,
"age": 0.0,
"candid": "1558254455015015000",
"n_det": 1,
},
"id": 916700,
"modified": "2021-04-08T06:17:56.980090",
"obj_id": "ZTF21aatjavc",
"origin": "au-public:hosted",
}]
Alert filters in Fritz¶
This section describes how to define alert stream filters within Fritz and provides some examples for reference.
Introduction¶
ZTF alerts are
generated at IPAC based on difference
imaging analysis and are distributed to the world at low latency via a Kafka alert stream.
Fritz’s Kowalski backend consumes this stream, persisting the alerts to a MongoDB database,
and supplementing them with other useful quantities such as Galactic coordinates, external catalog cross-matches,
machine learning scores etc. Next, Kowalski executes a series of user-defined filters on each new (“enhanced”)
incoming alert accessible to the filter. Users create filters on the SkyPortal frontend and they are executed
on the Kowalski backend. If an alert passes a filter, it is pushed up to SkyPortal and appears on a program’s
scanning page.
Note: for a detailed description of the ZTF alerts and their contents, please see here.
Implementation of Filters as MongoDB Aggregation Pipelines¶
Kowalski uses MongoDB, a document-based NoSQL database, on the backend.
For a very brief introduction into
MongoDB, we recommend watching MongoDB in 5 Minutes with Eliot Horowitz.If you are familiar with relational databases, you may want to check out the SQL to MongoDB Mapping Chart.
MongoDB database stores its data in “collections”. A collection holds one or more “documents”.
Documents are analogous to records or rows in a relational database table.
Each document has one or more “fields”; fields are similar to the columns in a relational database table.
MongoDB supports a rich query language, which we will make use of when working with the alert filters.
Alert filtering is implemented as a MongoDB aggregation pipeline that first “massages” the newly ingested alert data such that the user’s filter deals with enhanced “packets” containing, for example, longer photometry history (and not just the rolling 30-day window), cross-match data, and custom ML scores.
The MongoDB aggregation pipeline is a framework for data aggregation modeled on the concept of data processing pipelines. Documents enter a multi-stage pipeline that transforms the documents into aggregated results.
An aggregation pipeline is represented as a list of dictionaries, each corresponding to a processing stage/step:
[
{<stage_1>},
...
{<stage_N>}
]
Documents pass through the stages in sequence. Different stages can appear multiple times in a pipeline.
As of MongoDB version 4.2, there are 30 different types of aggregation pipeline stages, please see the official documentation for a detailed description.
To manipulate documents, each stage uses expressions, which can include field paths, literals, system variables, expression objects, and expression operators. Expressions can be nested.
Testing your filter¶
To ease the process of writing and debugging the filters, we have set up two live public MongoDB Atlas databases
in the cloud:
The first one contains a small curated set of ~300 sample public ZTF alerts originating from SNe, variable stars, AGN, and bogus detections. The auxiliary information is limited to the detection history present in the alert packets.
The second one contains ~120,000 public ZTF alerts from July 6, 2020. The auxiliary information contains a ~100-day history of detections (limited to reduce the test database size), cross-matches with external catalogs, and a few additional computed quantities.
We recommend to begin exploring filtering on the first database and then move onto the second one.
We will show how to use a tool called MongoDB Compass
(the full version is now free) to construct and debug aggregation pipelines (aka alert filters), using the two cloud
databases, that can be then plugged into Fritz.
Filters will be managed on a dedicated page on Fritz. A detailed description of the interface and its capabilities
(including, for example, filter versioning and diff’ing) will be covered elsewhere – please stay tuned.
MongoDB Compass¶
Download and install MongoDB Compass for your system from here.
The connection string to access the first sample public alert database:
mongodb://ztf:FritzZwicky@fritz-test-shard-00-00-uas9d.gcp.mongodb.net:27017,fritz-test-shard-00-01-uas9d.gcp.mongodb.net:27017,fritz-test-shard-00-02-uas9d.gcp.mongodb.net:27017/test?authSource=admin&replicaSet=fritz-test-shard-0&readPreference=primary&appname=MongoDB%20Compass&ssl=true
The connection string to access the second sample public alert database:
mongodb+srv://ztf:FritzZwicky@fritz-public-20200706.uas9d.gcp.mongodb.net/kowalski
Upon connection:
Select the
kowalskidatabaseSelect the
ZTF_alertscollectionGo to the Aggregations tab
Click the down arrow located to the left of the “Collation” button and select “New Pipeline From Text”
Copy paste the contents of the provided JSON files with the sample filters (see below)
Note that Compass shows the output of all stages interactively and also displays/explains any errors in the code, which is extremely helpful for debugging and experimentation.
By default, Compass’ aggregation pipeline builder works in Sample Mode, showing up to 20 documents that pass any given stage. You can change the default settings, including the default timeout by clicking the button with a little gear next to the “Auto Preview” toggle switch.

Note: the filter code that you will save on Fritz must be valid JSON, so, once you’re done with debugging, when exporting a filter in Compass, select “NODE” under “Export Pipeline To:”, copy-paste into a text editor, and then manually replace single quotes with double quotes. We also recommend using a text editor that can validate JSON.
“Upstream” aggregation pipeline stages¶
[OPTIONAL READ]
The upstream “massaging” mentioned above is performed by Fritz for each alert and includes:
Selecting the newly ingested alert from the
ZTF_alertscollection by itscandidRemoving the image cutouts to reduce traffic
Joining the alert by its
objectIdwith the corresponding entry in theZTF_alerts_auxcollection, which contains the cross-matches, ML scores, computed quantities, and archival photometry / detection history
The upstream stages also take care of the ACLs.
Fritz uses the following four stages:
The first
$matchstage selects the alert by its candid and ensures the ACLs are respected.
For example, for a program that has access to the partnership data:
{
"$match": {
"candid": 1282486310015015001,
"candidate.programid": {
"$in": [1, 2]
}
}
}
TIP: When debugging your filter with Compass, you may want to play around with this stage: try selecting an object
that must have passed the filter (when using the second database, that must have happened on July 6, 2020) by
specifying "objectId": "<ZTF object id>", or turning it off altogether to make Mongo look at all the alerts stored
in the sample database.
The image cutouts are stored per alert, but generally not needed for the filtering purposes. The
$projectstage removes them:
{
"$project": {
"cutoutScience": 0,
"cutoutTemplate": 0,
"cutoutDifference": 0
}
}
Using the
$lookupstage, the alert data are joined with auxiliary data stored in theZTF_alerts_auxcollection, which uses the alert’sobjectIds as unique document identifiers (_id- a concept in MongoDB similar to primary keys in SQL):
{
"$lookup": {
"from": "ZTF_alerts_aux",
"localField": "objectId",
"foreignField": "_id",
"as": "aux"
}
}
The final
$projectstage reshapes the joined data for convenience, selects the last 365 days of photometry history (which is done having practical considerations in mind and may be relaxed in the future), and applies ACLs to the detection history stored inprv_candidates:
{
"$project": {
"cross_matches": {
"$arrayElemAt": [
"$aux.cross_matches",
0
]
},
"prv_candidates": {
"$filter": {
"input": {
"$arrayElemAt": [
"$aux.prv_candidates",
0
]
},
"as": "item",
"cond": {
"$and": [
{
"$in": [
"$$item.programid",
[
1,
2,
3
]
]
},
{
"$lt": [
{
"$subtract": [
"$candidate.jd",
"$$item.jd"
]
},
365
]
}
]
}
}
},
"schemavsn": 1,
"publisher": 1,
"objectId": 1,
"candid": 1,
"candidate": 1,
"classifications": 1,
"coordinates": 1
}
}
The user-defined filter stages then operate on the “enhanced” packets that look like this:
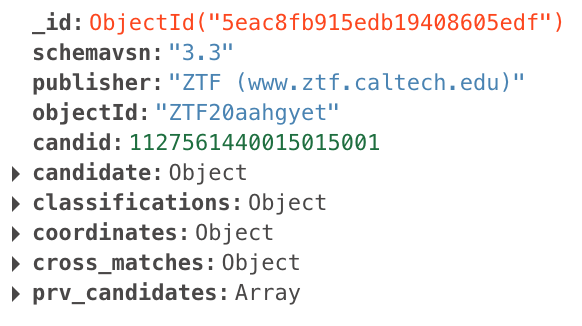
Fritz automatically prepends these stages to all user-defined filters. However, when constructing/debugging
filters in Compass, the users must take care of that – all the examples below come with the upstream stages prepended.
Limitations¶
$lookup, $unionWith, $out, and $merge stages are not allowed in the user-defined part of the filters.
Alert data augmentation¶
Fritz’s Kowalski backend augments the alert data with the following: [as of January 2021]
Galactic coordinates
Cross-matches with external catalogs:
2MASS_PSC (all matches within 2”)
AllWISE (all matches within 2”)
GALEX (all matches within 2”)
Gaia_DR2 (all matches within 2”)
Gaia_EDR3 (all matches within 2”)
Gaia_DR2_WD (all matches within 2”)
IPHAS_DR2 (all matches within 2”)
LAMOST_DR5_v3 (all matches within 2”)
PS1_DR1 (all matches within 2”)
PS1_STRM (all matches within 2”)
galaxy_redshifts_20200522 (all matches within 2”)
CLU_20190625 (“elliptical” matches with close galaxies using 3x their size)
For the detailed description of the available catalogs, see here
Machine learning scores:
braaiversiond6_m9– real/bogus classifieracai_hversiond1_dnn_20201130– phenomenological classifier, “hosted”acai_oversiond1_dnn_20201130– phenomenological classifier, “orphan”acai_nversiond1_dnn_20201130– phenomenological classifier, “nuclear”acai_vversiond1_dnn_20201130– phenomenological classifier, “variable star”acai_bversiond1_dnn_20201130– phenomenological classifier, “bogus”
ACAI¶
In November 2020, we deployed a set of new phenomenological deep learning classifiers called ACAI (Alert-Classifying AI; publication in prep.). The system consists of 5 binary classifiers:
acai_h– “hosted” – genuine transient in the vicinity of a “host” with (some) morphology, e.g. something one could call a galaxy; should catch SN, Novae etcacai_o– “orphan” – a genuine orphan transient, i.e. there are no identifiable “hosts” in its vicinity; catches asteroids and hostless (or with hosts that are too faint) transientsacai_n– “nuclear” – a genuine transient occurring in a galaxy/quasar nucleus; should catch AGN, TDEs, etc.acai_v– “variable star” – variable staracai_b– “bogus” – a new version of the real/bogus classifier; could be thought of as (1 - braai)
Each classifier takes as input 25 features from the candidate section of a ZTF alert packet and a stack of full-sized thumbnails (science/reference (template)/difference) and produces a score from 0 to 1:

All classifiers were trained on more than 200,000 diverse alerts covering a large part of the phase space. The classifiers, although potentially correlated in their output, act independently and so can be mixed and matched and applied alongside other alert features.
Filter examples¶
Let us explore some examples! The provided JSON files contain prepended upstream stages, but they are not shown in the text below for brevity.
Simple filter¶
Let us start with a very simplistic example:
The first user-defined $match
stage selects alerts with deep-learning-based real-bogus scores (candidate.drb) greater than 0.9999999
that don’t have any matches with the Gaia_DR2 catalog
(the zeroth element of the corresponding array does not exist)
{
"$match": {
"candidate.drb": {
"$gt": 0.9999999
},
"cross_matches.Gaia_DR2.0": {
"$exists": false
}
}
}
In case an alert passes the first stage, the output stage adds the annotations dictionary:
{
"$project": {
"_id": 0,
"candid": 1,
"objectId": 1,
"annotations.author": "dd",
"annotations.mean_rb": {
"$avg": "$prv_candidates.rb"
}
}
}
candid and objectId are kept here for the debugging purposes, to see which alerts pass your filter.
The output of the filter will look something like this:
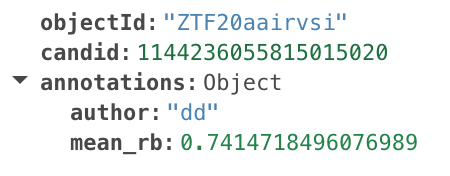
This alert will be posted to the candidates page with these annotations.
ACAI-hosted filter¶
Let us build a filter that primarily relies on the ACAI ML models to select transients that are confidently
classified as “hosted” (and nothing else), and are a positive subtraction (candidate.isdiffpos) and
not a known Solar system object (candidate.ssdistnr):
[
{
"$match": {
"classifications.acai_h": {"$gte": 0.8},
"classifications.acai_b": {"$lt": 0.1},
"classifications.acai_v": {"$lt": 0.1},
"classifications.acai_o": {"$lt": 0.4},
"classifications.acai_n": {"$lt": 0.4},
"candidate.ssdistnr": {"$lt": 0},
"candidate.isdiffpos": {"$in": [1, "1", true, "t"]}
}
},
{
"$project": {
"annotations.age": {"$round": [{"$subtract": ["$candidate.jd", "$candidate.jdstarthist"]}, 5]},
"annotations.n_det": "$candidate.ndethist",
"annotations.candid": {"$toString": "$candid"},
"annotations.acai_h": {"$round": ["$classifications.acai_h", 5]},
"annotations.acai_v": {"$round": ["$classifications.acai_v", 5]},
"annotations.acai_o": {"$round": ["$classifications.acai_o", 5]},
"annotations.acai_n": {"$round": ["$classifications.acai_n", 5]},
"annotations.acai_b": {"$round": ["$classifications.acai_b", 5]}
}
}
]
CLU filter¶
Now that we’ve looked at basic examples, let us explore a more complex case and build a filter for the Census of the Local Universe program.
As a reference, we will use the filter definition (as of July 10, 2020)
from the GROWTH marshal translated into python code (copy-pasted here with no alterations):
def clu_filter(current_observation):
filteron = False
annotations = {}
calccount = 10000
bright = False
nopointunderneath = True
mover = True
real = False
tdiff = (-99.0)
magdiff = (-99)
riserate = (-99)
decayrate = (-99)
hostgr = (-99)
hostri = (-99)
positivesubtraction = False
brightstar = False
highlum = False
deltajd = 0
prevcandidates = current_observation['prv_candidates']
m_now = current_observation['candidate']['magpsf']
m_app = current_observation['candidate']['magap']
t_now = current_observation['candidate']['jd']
fid_now = current_observation['candidate']['fid']
sgscore = current_observation['candidate']['sgscore1']
sgscore2 = current_observation['candidate']['sgscore2']
sgscore3 = current_observation['candidate']['sgscore3']
srmag = current_observation['candidate']['srmag1']
srmag2 = current_observation['candidate']['srmag2']
srmag3 = current_observation['candidate']['srmag3']
sgmag = current_observation['candidate']['sgmag1']
simag = current_observation['candidate']['simag1']
rbscore = current_observation['candidate']['rb']
magnr = current_observation['candidate']['magnr']
distnr = current_observation['candidate']['distnr']
distpsnr1 = current_observation['candidate']['distpsnr1']
distpsnr2 = current_observation['candidate']['distpsnr2']
distpsnr3 = current_observation['candidate']['distpsnr3']
scorr = current_observation['candidate']['scorr']
fwhm = current_observation['candidate']['fwhm']
elong = current_observation['candidate']['elong']
nbad = current_observation['candidate']['nbad']
chipsf = current_observation['candidate']['chipsf']
gal_lat = current_observation['candidate']['gal_lat']
jdstarthist = current_observation['candidate']['jdstarthist']
jdendhist = current_observation['candidate']['jdendhist']
ssdistnr = current_observation['candidate']['ssdistnr']
ssmagnr = current_observation['candidate']['ssmagnr']
drb = current_observation['candidate']['drb']
if (jdstarthist and jdendhist):
deltajd = jdendhist - jdstarthist
calccount -= 2
psfminap = m_now - m_app
bright = m_now < 99.0
if (current_observation['candidate']['isdiffpos'] and (current_observation['candidate']['isdiffpos'] == 't' or current_observation['candidate']['isdiffpos'] == '1')):
positivesubtraction = True
calccount -= 2
if (rbscore and rbscore > 0.3 and drb > 0.5 and fwhm > 0.5 and fwhm < 8 and nbad < 5 and (psfminap < 0.75 or psfminap > (-0.75))):
real = True
calccount -= 2
if (sgscore and distpsnr1 and sgscore > 0.76 and distpsnr1 < 2):
nopointunderneath = False
calccount -= 2
if ((distpsnr1 and srmag and distpsnr1 < 20 and srmag < 15.0 and srmag > 0 and sgscore > 0.49) or (distpsnr2 and srmag2 and distpsnr2 < 20 and srmag2 < 15.0 and srmag2 > 0 and sgscore2 > 0.49) or (distpsnr3 and srmag3 and distpsnr3 < 20 and srmag3 < 15.0 and srmag3 > 0 and sgscore3 > 0.49)):
brightstar = True
calccount -= 2
for candidate in prevcandidates:
calccount -= 2
if (candidate['jd'] and candidate['magpsf'] and candidate['fid'] and candidate['isdiffpos'] and (candidate['isdiffpos'] == 't' or candidate['isdiffpos'] == '1')):
dt = t_now - candidate['jd']
if (dt > 0.02 and candidate['magpsf'] < 99 and (ssdistnr > 2 or ssdistnr < (-0.5))):
mover = False
calccount -= 2
calccount -= 3
if calccount < 0:
break
prevcandidates = current_observation['prv_candidates']
m_now = current_observation['candidate']['magpsf']
m_max = current_observation['candidate']['magpsf']
m_min = current_observation['candidate']['magpsf']
t_now = current_observation['candidate']['jd']
t_max = current_observation['candidate']['jd']
t_min = current_observation['candidate']['jd']
fid_now = current_observation['candidate']['fid']
for candidate in prevcandidates:
calccount -= 2
if (candidate['jd'] and candidate['magpsf'] and candidate['fid'] and candidate['isdiffpos'] and (candidate['isdiffpos'] == 't' or candidate['isdiffpos'] == '1')):
if (candidate['fid'] and candidate['fid'] == fid_now):
if (m_now < 99 and candidate['magpsf'] < 99):
if (candidate['magpsf'] > m_max):
m_max = candidate['magpsf']
t_max = candidate['jd']
calccount -= 3
else:
if (candidate['magpsf'] < m_min):
m_min = candidate['magpsf']
t_min = candidate['jd']
calccount -= 3
calccount -= 2
calccount -= 3
calccount -= 2
calccount -= 2
if calccount < 0:
break
tdiff = t_max - t_min
magdiff = m_min - m_max
if (tdiff != 0):
if (tdiff > 0):
riserate = magdiff / tdiff
calccount -= 2
else:
decayrate = (-magdiff) / tdiff
calccount -= 2
calccount -= 3
if (ssdistnr < 2 and ssdistnr > (-0.5)):
mover = True
calccount -= 2
hostgr = sgmag - srmag
hostri = srmag - simag
annotations['FWHM'] = fwhm
annotations['host g-r'] = hostgr
annotations['host r-i'] = hostri
annotations['mag at max'] = m_max
annotations['time at max'] = t_max
annotations['min-mag'] = m_min
annotations['min-time'] = t_min
annotations['time difference'] = tdiff
annotations['mag diff'] = magdiff
annotations['rise rate'] = riserate
annotations['decay rate'] = decayrate
annotations['host ZTF ref PSF r-mag'] = magnr
annotations['PS1 psf r-mag'] = srmag
annotations['rb score'] = rbscore
annotations['sgscore1'] = sgscore
annotations['ZOGI scorr'] = scorr
annotations['distpsnr1'] = distpsnr1
annotations['distpsnr2'] = distpsnr2
annotations['distpsnr3'] = distpsnr3
annotations['magpsf'] = m_now
annotations['elongation'] = elong
annotations['magap_min_magpsf'] = psfminap
annotations['gal_lat'] = gal_lat
annotations['deltajd'] = deltajd
filteron = bright and nopointunderneath and ((not mover)) and real and positivesubtraction and ((not brightstar))
return filteron, annotations
The Fritz-implementation that can be loaded into Compass can be found here:
Let us explore it step-by-step and look at the individual user-defined stages, again omitting the upstream part.
Note: try turning off the first stage of the pipeline that is pre-configured to select alerts by objectId
for demo purposes. When working with the 20200706 database, try a different objectId, for example ZTF20abjoqjy.
The first $match stage checks whether
the alert has at least one match with the CLU catalog:
{
"$match": {
"cross_matches.CLU_20190625.0": {
"$exists": true
}
}
}
In the following $project
stage, we define the variables that will be used by the downstream stages.
Note how we are using the $subtract
operator for the deltajd and psfminap fields.
Also, the candidates_fid field concatenates the previous >3sigma detections in the same filter as fid_now
with the current photometric point, which we will use below.
For the full list of MongoDB’s aggregation pipeline operators, see here.
{
"$project": {
"_id": 0,
"candid": 1,
"objectId": 1,
"prv_candidates.jd": 1,
"prv_candidates.magpsf": 1,
"prv_candidates.fid": 1,
"prv_candidates.isdiffpos": 1,
"isdiffpos": "$candidate.isdiffpos",
"m_now": "$candidate.magpsf",
"m_app": "$candidate.magap",
"t_now": "$candidate.jd",
"fid_now": "$candidate.fid",
"sgscore": "$candidate.sgscore1",
"sgscore2": "$candidate.sgscore2",
"sgscore3": "$candidate.sgscore3",
"srmag": "$candidate.srmag1",
"srmag2": "$candidate.srmag2",
"srmag3": "$candidate.srmag3",
"sgmag": "$candidate.sgmag1",
"simag": "$candidate.simag1",
"rbscore": "$candidate.rb",
"drb": "$candidate.drb",
"magnr": "$candidate.magnr",
"distnr": "$candidate.distnr",
"distpsnr1": "$candidate.distpsnr1",
"distpsnr2": "$candidate.distpsnr2",
"distpsnr3": "$candidate.distpsnr3",
"scorr": "$candidate.scorr",
"fwhm": "$candidate.fwhm",
"elong": "$candidate.elong",
"nbad": "$candidate.nbad",
"chipsf": "$candidate.chipsf",
"gal_lat": "$coordinates.b",
"ssdistnr": "$candidate.ssdistnr",
"ssmagnr": "$candidate.ssmagnr",
"ssnamenr": "$candidate.ssnamenr",
"jdstarthist": "$candidate.jdstarthist",
"jdendhist": "$candidate.jdendhist",
"deltajd": {
"$subtract": [
"$candidate.jdendhist", "$candidate.jdstarthist"
]
},
"psfminap": {
"$subtract": [
"$candidate.magpsf", "$candidate.magap"
]
},
"candidates_fid": {
"$concatArrays": [
{
"$filter": {
"input": "$prv_candidates",
"as": "cand",
"cond": {
"$and": [
{
"$eq": [
"$$cand.fid", "$candidate.fid"
]
},
{
"$gt": [
"$$cand.magpsf", 0
]
},
{
"$lt": [
"$$cand.magpsf", 99
]
}
]
}
}
},
[
{
"jd": "$candidate.jd",
"magpsf": "$candidate.magpsf"
}
]
]
}
}
}
In the next stage, we define several boolean fields
(bright, positivesubtraction, real, nopointunderneath, brightstar, variablesource, rock, stationary),
pass several of the fields defined above (such as, e.g., m_now), and save indexes of the
maximum and minimum magnitudes in the candidates_fid array, which we will use in the next stage.
{
"$project": {
"t_now": 1,
"m_now": 1,
"fid_now": 1,
"sgscore": 1,
"drbscore": 1,
"magnr": 1,
"distnr": 1,
"scorr": 1,
"ssdistnr": 1,
"ssnamenr": 1,
"rbscore": 1,
"drb": 1,
"sgmag": 1,
"srmag": 1,
"simag": 1,
"distpsnr1": 1,
"distpsnr2": 1,
"distpsnr3": 1,
"fwhm": 1,
"elong": 1,
"gal_lat": 1,
"jdstarthist": 1,
"jdendhist": 1,
"psfminap": 1,
"candidates_fid": 1,
"m_max_index": {
"$indexOfArray": [
"$candidates_fid.magpsf", {
"$max": [
"$candidates_fid.magpsf"
]
}
]
},
"m_min_index": {
"$indexOfArray": [
"$candidates_fid.magpsf", {
"$min": [
"$candidates_fid.magpsf"
]
}
]
},
"bright": {
"$lt": [
"$m_now", 99.0
]
},
"positivesubtraction": {
"$in": [
"$isdiffpos",
[
1, "1", "t", true
]
]
},
"real": {
"$and": [
{
"$gt": [
"$rbscore", 0.3
]
}, {
"$gt": [
"$drb", 0.5
]
}, {
"$gt": [
"$fwhm", 0.5
]
}, {
"$lt": [
"$fwhm", 8
]
}, {
"$lt": [
"$nbad", 5
]
}, {
"$lt": [
{
"$abs": "$psfminap"
}, 0.75
]
}
]
},
"nopointunderneath": {
"$not": [
{
"$and": [
{
"$gt": [
"$sgscore", 0.76
]
}, {
"$lt": [
"$distpsnr1", 2
]
}
]
}
]
},
"brightstar": {
"$or": [
{
"$and": [
{
"$lt": [
"$distpsnr1", 20
]
}, {
"$lt": [
"$srmag", 15
]
}, {
"$gt": [
"$srmag", 0
]
}, {
"$gt": [
"$sgscore", 0.49
]
}
]
}, {
"$and": [
{
"$lt": [
"$distpsnr2", 20
]
}, {
"$lt": [
"$srmag2", 15
]
}, {
"$gt": [
"$srmag2", 0
]
}, {
"$gt": [
"$sgscore2", 0.49
]
}
]
}, {
"$and": [
{
"$lt": [
"$distpsnr3", 20
]
}, {
"$lt": [
"$srmag3", 15
]
}, {
"$gt": [
"$srmag3", 0
]
}, {
"$gt": [
"$sgscore3", 0.49
]
}
]
}, {
"$and": [
{
"$eq": [
"$sgscore", 0.5
]
}, {
"$lt": [
"$distpsnr1", 0.5
]
}, {
"$or": [
{
"$lt": [
"$sgmag", 17
]
}, {
"$lt": [
"$srmag", 17
]
}, {
"$lt": [
"$simag", 17
]
}
]
}
]
}
]
},
"variablesource": {
"$or": [
{
"$and": [
{
"$lt": [
"$distnr", 0.4
]
}, {
"$lt": [
"$magnr", 19
]
}, {
"$gt": [
"$age", 90
]
}
]
}, {
"$and": [
{
"$lt": [
"$distnr", 0.8
]
}, {
"$lt": [
"$magnr", 17
]
}, {
"$gt": [
"$age", 90
]
}
]
}, {
"$and": [
{
"$lt": [
"$distnr", 1.2
]
}, {
"$lt": [
"$magnr", 15
]
}, {
"$gt": [
"$age", 90
]
}
]
}
]
},
"rock": {
"$and": [
{
"$gte": [
"$ssdistnr", 0
]
}, {
"$lt": [
"$ssdistnr", 12
]
}, {
"$lt": [
{
"$abs": "$ssmagnr"
}, 20
]
}
]
},
"stationary": {
"$anyElementTrue": {
"$map": {
"input": "$prv_candidates",
"as": "cand",
"in": {
"$and": [
{
"$gt": [
{
"$abs": {
"$subtract": [
"$t_now", "$$cand.jd"
]
}
}, 0.02
]
}, {
"$lt": [
"$$cand.magpsf", 99
]
}, {
"$in": [
"$$cand.isdiffpos", [
1, "1", true, "t"
]
]
}, {
"$or": [
{
"$lt": [
"$ssdistnr", -0.5
]
}, {
"$gt": [
"$ssdistnr", 2
]
}
]
}
]
}
}
}
}
}
}
Let us take a closer look at a few of the fields defined above:
In the definition of
m_max_index, the$indexOfArrayoperator returns the index of the maximum value of themagpsffield in the detection history:
$candidates_fid.magpsfreturns an array containing the filteredmagpsfsthe
$maxoperator takes the maximum of that array
In the definition of
stationary, we evaluate a boolean statement (expressed using the$andoperator) on each entry in the full detection history stored in theprv_candidatesarray using the$mapoperator.
In the following $project stage, we compute the maximum and minimum magnitudes
for the source and the corresponding time stamps using the m_max_index and m_min_index values computed above:
{
"$project": {
"m_max": {
"$arrayElemAt": [
"$candidates_fid.magpsf", "$m_max_index"
]
},
"m_min": {
"$arrayElemAt": [
"$candidates_fid.magpsf", "$m_min_index"
]
},
"t_max": {
"$arrayElemAt": [
"$candidates_fid.jd", "$m_max_index"
]
},
"t_min": {
"$arrayElemAt": [
"$candidates_fid.jd", "$m_min_index"
]
},
"t_now": 1,
"m_now": 1,
"fid_now": 1,
"sgscore": 1,
"drbscore": 1,
"magnr": 1,
"distnr": 1,
"scorr": 1,
"gal_lat": 1,
"ssdistnr": 1,
"ssnamenr": 1,
"rbscore": 1,
"drb": 1,
"sgmag": 1,
"srmag": 1,
"simag": 1,
"distpsnr1": 1,
"distpsnr2": 1,
"distpsnr3": 1,
"fwhm": 1,
"elong": 1,
"jdstarthist": 1,
"jdendhist": 1,
"psfminap": 1,
"bright": 1,
"positivesubtraction": 1,
"real": 1,
"nopointunderneath": 1,
"brightstar": 1,
"variablesource": 1,
"rock": 1,
"stationary": 1
}
}
The alert is selected or rejected by our filter with a
$match
stage based on the values of the boolean fields defined above:
{
"$match": {
"bright": true,
"nopointunderneath": true,
"positivesubtraction": true,
"real": true,
"stationary": true,
"brightstar": false,
"rock": false
}
}
The final stage adds annotations to a passing alert:
{
"$project": {
"annotations.FWHM": "$fwhm",
"annotations.drb": "$drb",
"annotations.host g-r": {
"$subtract": [
"$sgmag", "$srmag"
]
},
"annotations.host r-i": {
"$subtract": [
"$srmag", "$simag"
]
},
"annotations.mag at max": "$m_max",
"annotations.time at max": "$t_max",
"annotations.min-mag": "$m_min",
"annotations.min-time": "$t_min",
"annotations.time difference": {
"$subtract": [
"$t_max", "$t_min"
]
},
"annotations.mag diff": {
"$subtract": [
"$m_min", "$m_max"
]
},
"annotations.rise rate": {
"$cond": {
"if": {
"$gt": [
{
"$subtract": [
"$t_max", "$t_min"
]
}, 0
]
},
"then": {
"$divide": [
{
"$subtract": [
"$m_min", "$m_max"
]
}, {
"$subtract": [
"$t_max", "$t_min"
]
}
]
},
"else": null
}
},
"annotations.decay rate": {
"$cond": {
"if": {
"$lt": [
{
"$subtract": [
"$t_max", "$t_min"
]
}, 0
]
},
"then": {
"$divide": [
{
"$subtract": [
"$m_max", "$m_min"
]
}, {
"$subtract": [
"$t_max", "$t_min"
]
}
]
},
"else": null
}
},
"annotations.host ZTF ref PSF r-mag": "$magnr",
"annotations.PS1 psf r-mag": "$srmag",
"annotations.rb score": "$rbscore",
"annotations.sgscore1": "$sgscore",
"annotations.ZOGI scorr": "$scorr",
"annotations.distpsnr1": "$distpsnr1",
"annotations.distpsnr2": "$distpsnr2",
"annotations.distpsnr3": "$distpsnr3",
"annotations.magpsf": "$m_now",
"annotations.elongation": "$elong",
"annotations.magap_min_magpsf": "$psfminap",
"annotations.gal_lat": "$gal_lat",
"annotations.deltajd": {
"$subtract": [
"$jdendhist", "$jdstarthist"
]
}
}
}
BTS/RCF program (simplified) filter¶
As another example, below you will find a simplified version of the Bright Transient Survey filter (the Redshift Completeness Factor program)
[
/* UPSTREAM STAGES */
// For this example, select alerts by objectId. In practice, alert is selected by candid
{
"$match": {
"objectId": "ZTF20aacbyec",
"candidate.programid": {
"$in": [
1
]
}
}
},
{
"$project": {
"cutoutScience": 0,
"cutoutTemplate": 0,
"cutoutDifference": 0
}
},
{
"$lookup": {
"from": "ZTF_alerts_aux",
"localField": "objectId",
"foreignField": "_id",
"as": "aux"
}
},
{
"$project": {
"cross_matches": {
"$arrayElemAt": [
"$aux.cross_matches", 0
]
},
"prv_candidates": {
"$filter": {
"input": {
"$arrayElemAt": [
"$aux.prv_candidates", 0
]
},
"as": "item",
"cond": {
"$in": [
"$$item.programid", [
1
]
]
}
}
},
"schemavsn": 1,
"publisher": 1,
"objectId": 1,
"candid": 1,
"candidate": 1,
"classifications": 1,
"coordinates": 1
}
},
/* USER-DEFINED PART */
{
"$project": {
"_id": 0,
"candid": 1,
"objectId": 1,
"prv_candidates.jd": 1,
"prv_candidates.magpsf": 1,
"prv_candidates.fid": 1,
"prv_candidates.isdiffpos": 1,
"isdiffpos": "$candidate.isdiffpos",
"m_now": "$candidate.magpsf",
"m_app": "$candidate.magap",
"t_now": "$candidate.jd",
"fid_now": "$candidate.fid",
"sgscore": "$candidate.sgscore1",
"sgscore2": "$candidate.sgscore2",
"sgscore3": "$candidate.sgscore3",
"srmag": "$candidate.srmag1",
"srmag2": "$candidate.srmag2",
"srmag3": "$candidate.srmag3",
"sgmag": "$candidate.sgmag1",
"simag": "$candidate.simag1",
"drbscore": "$candidate.drb",
"magnr": "$candidate.magnr",
"distnr": "$candidate.distnr",
"distpsnr1": "$candidate.distpsnr1",
"distpsnr2": "$candidate.distpsnr2",
"distpsnr3": "$candidate.distpsnr3",
"scorr": "$candidate.scorr",
"fwhm": "$candidate.fwhm",
"elong": "$candidate.elong",
"nbad": "$candidate.nbad",
"chipsf": "$candidate.chipsf",
"gal_lat": "$coordinates.b",
"ssdistnr": "$candidate.ssdistnr",
"ssmagnr": "$candidate.ssmagnr",
"ssnamenr": "$candidate.ssnamenr",
"t_start": "$candidate.jdstarthist",
"age": {
"$subtract": [
"$candidate.jd", "$candidate.jdstarthist"
]
}
}
},
{
"$project": {
"objectId": 1,
"t_now": 1,
"m_now": 1,
"fid_now": 1,
"sgscore": 1,
"drbscore": 1,
"magnr": 1,
"distnr": 1,
"scorr": 1,
"gal_lat": 1,
"ssdistnr": 1,
"ssnamenr": 1,
"age": 1,
"peakmag": {
"$min": [
{
"$map": {
"input": "$prv_candidates",
"as": "cand",
"in": {
"$cond": [
{
"$eq": [
"$$cand.fid", "$fid_now"
]
}, "$$cand.magpsf", null
]
}
}
}, "$m_now"
]
},
"bright": {
"$or": [
{
"$lt": [
"$m_now", 19.0
]
}, {
"$map": {
"input": "$prv_candidates",
"as": "cand",
"in": {
"$and": [
{
"$lt": [
{
"$abs": {
"$subtract": [
"$t_now", "$$cand.jd"
]
}
}, 0.75
]
}, {
"$in": [
"$isdiffpos", [
1, "1", "t", true
]
]
}, {
"$gt": [
"$$cand.magpsf", 0
]
}, {
"$lt": [
"$$cand.magpsf", 19
]
}
]
}
}
}
]
},
"latitude": {
"$gte": [
{
"$abs": "$gal_lat"
}, 7
]
},
"positivesubtraction": {
"$in": [
"$isdiffpos", [
1, "1", "t", true
]
]
},
"real": {
"$gt": [
"$drbscore", 0.5
]
},
"nopointunderneath": {
"$not": [
{
"$and": [
{
"$gt": [
"$sgscore", 0.76
]
}, {
"$lt": [
"$distpsnr1", 2
]
}
]
}
]
},
"brightstar": {
"$or": [
{
"$and": [
{
"$lt": [
"$distpsnr1", 20
]
}, {
"$lt": [
"$srmag", 15
]
}, {
"$gt": [
"$srmag", 0
]
}, {
"$gt": [
"$sgscore", 0.49
]
}
]
}, {
"$and": [
{
"$lt": [
"$distpsnr2", 20
]
}, {
"$lt": [
"$srmag2", 15
]
}, {
"$gt": [
"$srmag2", 0
]
}, {
"$gt": [
"$sgscore2", 0.49
]
}
]
}, {
"$and": [
{
"$lt": [
"$distpsnr3", 20
]
}, {
"$lt": [
"$srmag3", 15
]
}, {
"$gt": [
"$srmag3", 0
]
}, {
"$gt": [
"$sgscore3", 0.49
]
}
]
}, {
"$and": [
{
"$eq": [
"$sgscore", 0.5
]
}, {
"$lt": [
"$distpsnr1", 0.5
]
}, {
"$or": [
{
"$lt": [
"$sgmag", 17
]
}, {
"$lt": [
"$srmag", 17
]
}, {
"$lt": [
"$simag", 17
]
}
]
}
]
}
]
},
"variablesource": {
"$or": [
{
"$and": [
{
"$lt": [
"$distnr", 0.4
]
}, {
"$lt": [
"$magnr", 19
]
}, {
"$gt": [
"$age", 90
]
}
]
}, {
"$and": [
{
"$lt": [
"$distnr", 0.8
]
}, {
"$lt": [
"$magnr", 17
]
}, {
"$gt": [
"$age", 90
]
}
]
}, {
"$and": [
{
"$lt": [
"$distnr", 1.2
]
}, {
"$lt": [
"$magnr", 15
]
}, {
"$gt": [
"$age", 90
]
}
]
}
]
},
"rock": {
"$and": [
{
"$gte": [
"$ssdistnr", 0
]
}, {
"$lt": [
"$ssdistnr", 12
]
}, {
"$lt": [
{
"$abs": "$ssmagnr"
}, 20
]
}
]
},
"stationary": {
"$anyElementTrue": {
"$map": {
"input": "$prv_candidates",
"as": "cand",
"in": {
"$and": [
{
"$gt": [
{
"$abs": {
"$subtract": [
"$t_now", "$$cand.jd"
]
}
}, 0.02
]
}, {
"$lt": [
"$$cand.magpsf", 99
]
}, {
"$in": [
"$$cand.isdiffpos", [
1, "1", true, "t"
]
]
}
]
}
}
}
}
}
},
{
"$match": {
"latitude": true,
"bright": true,
"nopointunderneath": true,
"positivesubtraction": true,
"real": true,
"stationary": true,
"brightstar": false,
"rock": false
}
},
{
"$project": {
"objectId": 1,
"annotations.jd": "$t_now",
"annotations.magnitude": "$m_now",
"annotations.sgscore": "$sgscore",
"annotations.peakmag": "$peakmag",
"annotations.atpeak": {
"$eq": [
"$m_now", "$peakmag"
]
},
"annotations.age": "$age",
"annotations.drb": "$drbscore"
}
}
]
BTS/RCF program full filter¶
Here you will find the full version (as of December 2020) of the Bright Transient Survey filter (the Redshift Completeness Factor program)
Fast Transients program filter¶
In this example, we will see how to adapt the Fast Transients’ program filter that has been using Kowalski for the initial search.
In the python code snippet below, courtesy of Anna Ho and Yuhan Yao, a coarse search is run using Kowalski and then a
number of logical expressions would be evaluated on the query result:
from astropy.time import Time
import numpy as np
from penquins import Kowalski
# Set search window
obst = obs['UT_START'].values
start = Time(obst[0], format='isot').jd - 0.02
end = Time(obst[-1], format='isot').jd + 0.02
# gt is greater than, lt is lower than
q = {"query_type": "find",
"query": {
"catalog": "ZTF_alerts",
"filter": {
'candidate.jd': {'$gt': start, '$lt': end},
'candidate.magpsf': {'$lt': 20},
'candidate.isdiffpos': {'$in': ['1', 't']},
'candidate.programid': {'$gt': 0},
'candidate.ssdistnr': {'$lt': -1},
'candidate.drb': {'$gt': 0.65}, # That gives a 1.7% FNR and FPR.
},
"projection": {
"objectId": 1,
"candidate.distpsnr1": 1,
"candidate.sgscore1": 1,
"candidate.srmag1": 1,
"candidate.sgmag1": 1,
"candidate.simag1": 1,
"candidate.szmag1": 1,
}
}
}
kowalski = Kowalski()
query_result = kowalski.query(query=q)
out = query_result['data']
names_all = np.array([val['objectId'] for val in out])
names = np.unique(names_all)
print(f"There are {len(names)} unique cands from this initial filter.")
dist = np.array([val['candidate']['distpsnr1'] for val in out])
sg = np.array([val['candidate']['sgscore1'] for val in out])
rmag = np.array([val['candidate']['srmag1'] for val in out])
gmag = np.array([val['candidate']['sgmag1'] for val in out])
imag = np.array([val['candidate']['simag1'] for val in out])
zmag = np.array([val['candidate']['szmag1'] for val in out])
pointunderneath = np.zeros(len(names_all), dtype=bool)
crit = np.logical_and.reduce((dist>0, dist<2, sg>0.76))
pointunderneath[crit] = True
crit = np.logical_and(
np.logical_and.reduce((sg==0.5, dist>0, dist<0.5)),
np.logical_or.reduce((
np.logical_and(rmag>0,rmag<17),
np.logical_and(gmag>0,gmag<17),
np.logical_and(imag>0,imag<17),
np.logical_and(zmag>0,zmag<17),
)))
pointunderneath[crit] = True
crit = np.logical_or.reduce((
np.logical_and.reduce((rmag > 0, rmag < 12, sg > 0.49, dist < 20)),
np.logical_and.reduce((rmag > 0, rmag < 15, sg > 0.49, dist < 5)),
np.logical_and.reduce((rmag > 0, rmag < 15, sg > 0.8, dist < 20)),
np.logical_and.reduce((gmag > 0, gmag < 12, sg > 0.49, dist < 20)),
np.logical_and.reduce((gmag > 0, gmag < 15, sg > 0.8, dist < 20)),
np.logical_and.reduce((imag > 0, imag < 12, sg > 0.49, dist < 20)),
np.logical_and.reduce((imag > 0, imag < 11.5, sg > 0.49, dist < 20)),
np.logical_and.reduce((imag > 0, imag < 14.5, sg > 0.49, dist < 5)),
np.logical_and.reduce((imag > 0, imag < 15, sg > 0.8, dist < 20)),
np.logical_and.reduce((zmag > 0, zmag < 11.5, sg > 0.49, dist < 10)),
np.logical_and.reduce((zmag > 0, zmag < 14.0, sg > 0.49, dist < 2.5)),
np.logical_and.reduce((zmag > 0, zmag < 15, sg > 0.8, dist < 20))))
pointunderneath[crit] = True
names_no_star = np.unique(names_all[pointunderneath==False])
print(f"There are {len(names_no_star)} that are not stars.")
passed = np.array(names_no_star)
As we have seen above, we can build these kinds of logical expressions right into our filter:
[
{
"$project": {
"cutoutScience": 0,
"cutoutTemplate": 0,
"cutoutDifference": 0
}
},
{
"$lookup": {
"from": "ZTF_alerts_aux",
"localField": "objectId",
"foreignField": "_id",
"as": "aux"
}
},
{
"$project": {
"cross_matches": {
"$arrayElemAt": [
"$aux.cross_matches", 0
]
},
"prv_candidates": {
"$filter": {
"input": {
"$arrayElemAt": [
"$aux.prv_candidates", 0
]
},
"as": "item",
"cond": {
"$and": [
{
"$in": [
"$$item.programid", [
1, 2, 3
]
]
}, {
"$lt": [
{
"$subtract": [
"$candidate.jd", "$$item.jd"
]
}, 100
]
}
]
}
}
},
"schemavsn": 1,
"publisher": 1,
"objectId": 1,
"candid": 1,
"candidate": 1,
"classifications": 1,
"coordinates": 1
}
},
{
"$match": {
"candidate.magpsf": {
"$lt": 20
},
"candidate.isdiffpos": {
"$in": [
"1", "t"
]
},
"candidate.programid": {
"$gt": 0
},
"candidate.ssdistnr": {
"$lt": -1
},
"candidate.drb": {
"$gt": 0.65
}
}
},
{
"$project": {
"objectId": 1,
"dist": "$candidate.distpsnr1",
"sg": "$candidate.sgscore1",
"rmag": "$candidate.srmag1",
"gmag": "$candidate.sgmag1",
"imag": "$candidate.simag1",
"zmag": "$candidate.szmag1"
}
},
{
"$project": {
"_id": 0,
"objectId": 1,
"pointunderneath": {
"$or": [
{
"$and": [
{
"$gt": [
"$dist", 0
]
}, {
"$lt": [
"$dist", 2
]
}, {
"$gt": [
"sg", 0.76
]
}
]
}, {
"$and": [
{
"$and": [
{
"$gt": [
"$dist", 0
]
}, {
"$lt": [
"$dist", 0.5
]
}, {
"$eq": [
"sg", 0.5
]
}
]
}, {
"$or": [
{
"$and": [
{
"$gt": [
"$rmag", 0
]
}, {
"$lt": [
"$rmag", 17
]
}
]
}, {
"$and": [
{
"$gt": [
"$gmag", 0
]
}, {
"$lt": [
"$gmag", 17
]
}
]
}, {
"$and": [
{
"$gt": [
"$imag", 0
]
}, {
"$lt": [
"$imag", 17
]
}
]
}, {
"$and": [
{
"$gt": [
"$zmag", 0
]
}, {
"$lt": [
"$zmag", 17
]
}
]
}
]
}
]
}, {
"$or": [
{
"$and": [
{
"$gt": [
"$rmag", 0
]
}, {
"$lt": [
"$rmag", 12
]
}, {
"$gt": [
"sg", 0.49
]
}, {
"$lt": [
"$dist", 20
]
}
]
}, {
"$and": [
{
"$gt": [
"$rmag", 0
]
}, {
"$lt": [
"$rmag", 15
]
}, {
"$gt": [
"sg", 0.49
]
}, {
"$lt": [
"$dist", 5
]
}
]
}, {
"$and": [
{
"$gt": [
"$rmag", 0
]
}, {
"$lt": [
"$rmag", 15
]
}, {
"$gt": [
"sg", 0.8
]
}, {
"$lt": [
"$dist", 20
]
}
]
}, {
"$and": [
{
"$gt": [
"$gmag", 0
]
}, {
"$lt": [
"$gmag", 12
]
}, {
"$gt": [
"sg", 0.49
]
}, {
"$lt": [
"$dist", 20
]
}
]
}, {
"$and": [
{
"$gt": [
"$gmag", 0
]
}, {
"$lt": [
"$gmag", 12
]
}, {
"$gt": [
"sg", 0.8
]
}, {
"$lt": [
"$dist", 20
]
}
]
}, {
"$and": [
{
"$gt": [
"$imag", 0
]
}, {
"$lt": [
"$imag", 12
]
}, {
"$gt": [
"sg", 0.49
]
}, {
"$lt": [
"$dist", 20
]
}
]
}, {
"$and": [
{
"$gt": [
"$imag", 0
]
}, {
"$lt": [
"$imag", 14.5
]
}, {
"$gt": [
"sg", 0.49
]
}, {
"$lt": [
"$dist", 5
]
}
]
}, {
"$and": [
{
"$gt": [
"$imag", 0
]
}, {
"$lt": [
"$imag", 15
]
}, {
"$gt": [
"sg", 0.8
]
}, {
"$lt": [
"$dist", 20
]
}
]
}, {
"$and": [
{
"$gt": [
"$zmag", 0
]
}, {
"$lt": [
"$zmag", 11.5
]
}, {
"$gt": [
"sg", 0.49
]
}, {
"$lt": [
"$dist", 10
]
}
]
}, {
"$and": [
{
"$gt": [
"$zmag", 0
]
}, {
"$lt": [
"$zmag", 14
]
}, {
"$gt": [
"sg", 0.49
]
}, {
"$lt": [
"$dist", 2.5
]
}
]
}, {
"$and": [
{
"$gt": [
"$zmag", 0
]
}, {
"$lt": [
"$zmag", 15
]
}, {
"$gt": [
"sg", 0.8
]
}, {
"$lt": [
"$dist", 20
]
}
]
}
]
}
]
}
}
},
{
"$match": {
"pointunderneath": false
}
},
{
"$project": {
"candid": 1,
"objectId": 1,
"annotations.comment": "fast!"
}
}
]
Watch lists¶
Fritz’s alert stream filters allow implementing watch lists for sets of objects of interest.
In the example below, we define two named sky positions of interest. The coordinates are given in degrees. Any alert from an object within 2 arcseconds from these positions will pass this filter.
[
// This is the only stage you would need to modify to set up your own watch list:
{
"$project": {
"_id": 0,
"watchlist": [
{
"name": "my_favorite_spot",
"ra": 134.0779551,
"dec": -0.4421303
},
{
"name": "my_star",
"ra": 14.5551,
"dec": 37.4451303
}
],
"max_distance_arcsec": {
"$literal": 2.0
},
"ra": {
"$degreesToRadians": "$candidate.ra"
},
"dec": {
"$degreesToRadians": "$candidate.dec"
}
}
},
// In this stage, we prepare the quantities that will be needed for the spherical distance computations
{
"$project": {
"watchlist": 1,
"max_distance_arcsec": 1,
"dec": 1,
"dra_dec": {
"$zip": {
"inputs": [
{
"$map": {
"input": "$watchlist",
"as": "object",
"in": {
"$subtract": [
"$ra", {
"$degreesToRadians": "$$object.ra"
}
]
}
}
}, {
"$map": {
"input": "$watchlist",
"as": "object",
"in": {
"$degreesToRadians": "$$object.dec"
}
}
}
]
}
}
}
},
// In this stage, we perform an accurate spherical distance computation
// for all objects in the watch list and convert the results to arcseconds:
{
"$project": {
"watchlist": 1,
"max_distance_arcsec": 1,
"distances_arcsec": {
"$map": {
"input": "$dra_dec",
"as": "s",
"in": {
"$multiply": [
3600.0, {
"$radiansToDegrees": {
"$atan2": [
{
"$sqrt": {
"$add": [
{
"$pow": [
{
"$multiply": [
{
"$cos": "$dec"
}, {
"$sin": {
"$arrayElemAt": [
"$$s", 0
]
}
}
]
}, 2
]
}, {
"$pow": [
{
"$subtract": [
{
"$multiply": [
{
"$cos": {
"$arrayElemAt": [
"$$s", 1
]
}
}, {
"$sin": "$dec"
}
]
}, {
"$multiply": [
{
"$sin": {
"$arrayElemAt": [
"$$s", 1
]
}
}, {
"$cos": "$dec"
}, {
"$cos": {
"$arrayElemAt": [
"$$s", 0
]
}
}
]
}
]
}, 2
]
}
]
}
}, {
"$add": [
{
"$multiply": [
{
"$sin": {
"$arrayElemAt": [
"$$s", 1
]
}
}, {
"$sin": "$dec"
}
]
}, {
"$multiply": [
{
"$cos": {
"$arrayElemAt": [
"$$s", 1
]
}
}, {
"$cos": "$dec"
}, {
"$cos": {
"$arrayElemAt": [
"$$s", 0
]
}
}
]
}
]
}
]
}
}
]
}
}
}
}
},
// Here, we look for objects that fall within max_distance_arcsec:
{
"$project": {
"watchlist": 1,
"distances_arcsec": 1,
"nearest_index": {
"$indexOfArray": [
"$distances_arcsec", {
"$min": "$distances_arcsec"
}
]
},
"bingo": {
"$filter": {
"input": {
"$zip": {
"inputs": [
"$watchlist", "$distances_arcsec"
]
}
},
"as": "item",
"cond": {
"$lte": [
{
"$arrayElemAt": [
"$$item", 1
]
}, "$max_distance_arcsec"
]
}
}
}
}
},
// We declare victory if such objects exist:
{
"$match": {
"bingo.0": {
"$exists": true
}
}
},
// Finally, we are adding some useful annotations:
{
"$project": {
"annotations.nearest_name": {
"$arrayElemAt": [
"$watchlist.name", "$nearest_index"
]
},
"annotations.nearest_distance_arcsec": {
"$round": [
{
"$arrayElemAt": [
"$distances_arcsec", "$nearest_index"
]
}, 3
]
},
"annotations.matches": {
"$map": {
"input": "$bingo",
"as": "item",
"in": {
"$arrayElemAt": [
"$$item", 0
]
}
}
},
"annotations.distances_arcsec": {
"$map": {
"input": "$bingo",
"as": "item",
"in": {
"$round": [
{
"$arrayElemAt": [
"$$item", 1
]
}, 3
]
}
}
}
}
}
]
Tips and tricks¶
prv_candidates array sorting¶
The prv_candidates are stored as a set and are thus not sorted. To do that, you may use the following stages
immediately after the default Fritz’s upstream stages:
[
{
'$project': {
'prv_candidates': {
'$concatArrays': [
'$prv_candidates', [
{
'fid': '$candidate.fid',
'jd': '$candidate.jd',
'magpsf': '$candidate.magpsf'
}
]
]
}
}
},
{
'$unwind': {
'path': '$prv_candidates'
}
},
{
'$sort': {
'prv_candidates.jd': 1
}
},
{
'$group': {
'_id': '$_id',
'prv_candidates': {
'$push': '$prv_candidates'
}
}
}
]
These will:
Squeeze in the current candidate into
prv_candidates(as it may or may not be there already)Unwind the
prv_candidatesarray into a bunch of documentsSort them by
jdGroup back into a single document with the
prv_candidatesin sorted order
Feedback¶
Please start by searching existing issues here. If the issue has already been reported, feel free to add to the existing discussion or to propose solutions.
If you found an as-yet-unreported problem, or you have a feature request, please create a new issue. The team will triage issues so that, within a few hours, you should see a label appear that indicates its development priority.